Giotto¶
Spatial transcriptomic and proteomic technologies have provided new opportunities to investigate cells in their native microenvironment. Here we present Giotto, a comprehensive and open-source toolbox for spatial data analysis and visualization. The analysis module provides end-to-end analysis by implementing a wide range of algorithms for characterizing tissue composition, spatial expression patterns, and cellular interactions. Furthermore, single-cell RNAseq data can be integrated for spatial cell-type enrichment analysis. The visualization module allows users to interactively visualize analysis outputs and imaging features. To demonstrate its general applicability, we apply Giotto to a wide range of datasets encompassing diverse technologies and platforms.
Example Functionalities¶
Analysis and visualization of large-scale spatial transcriptomic and proteomic datasets.
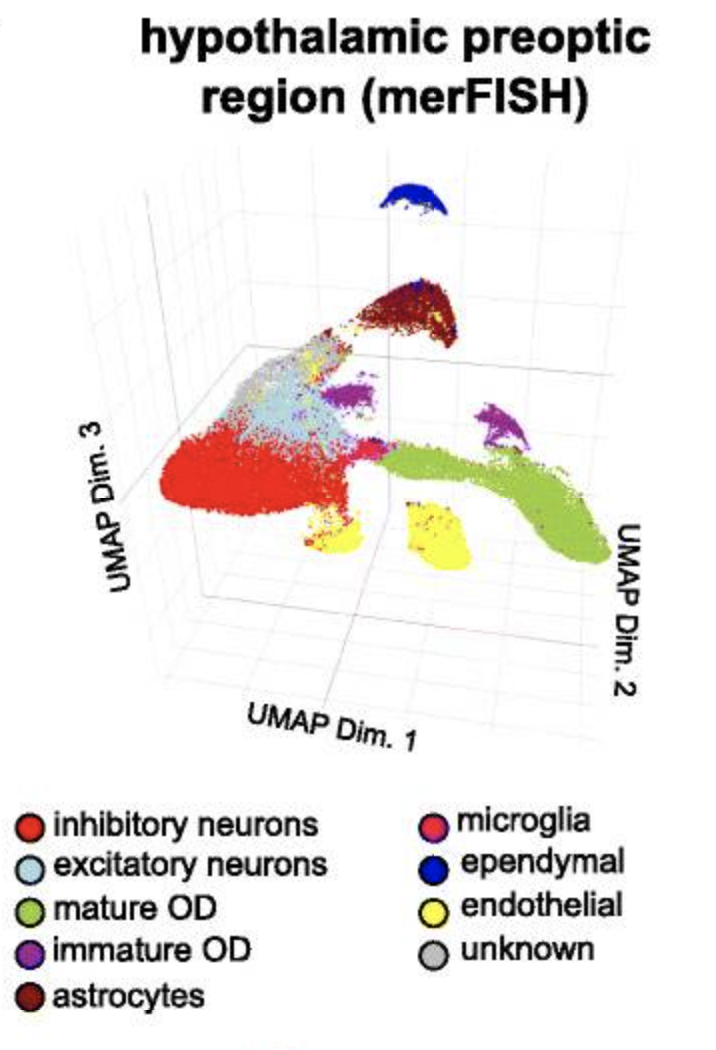
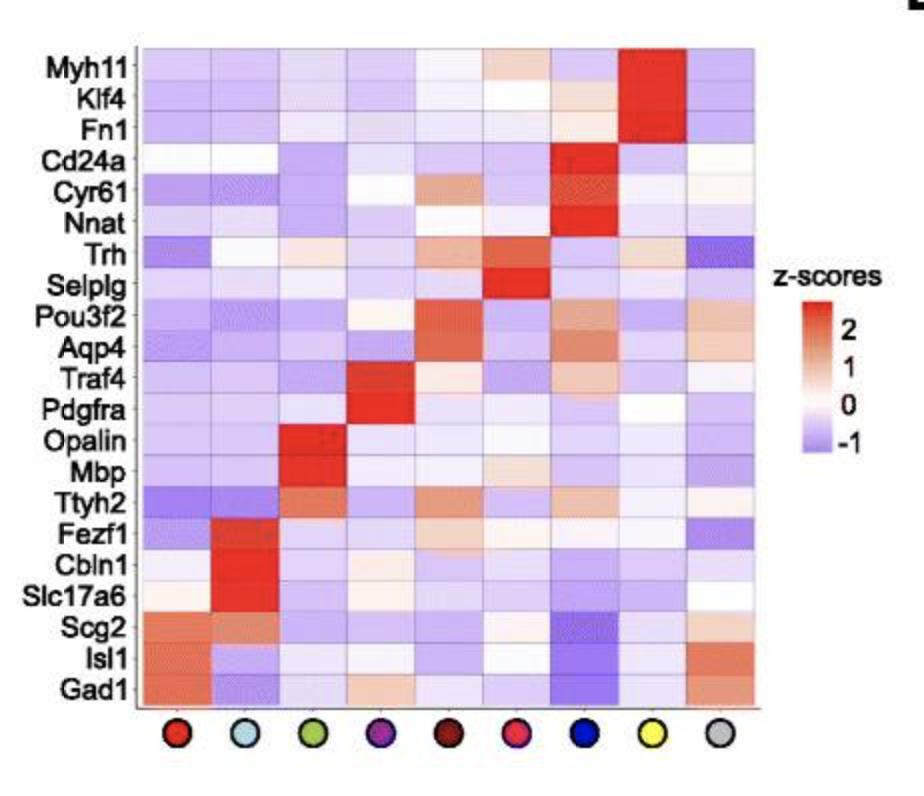
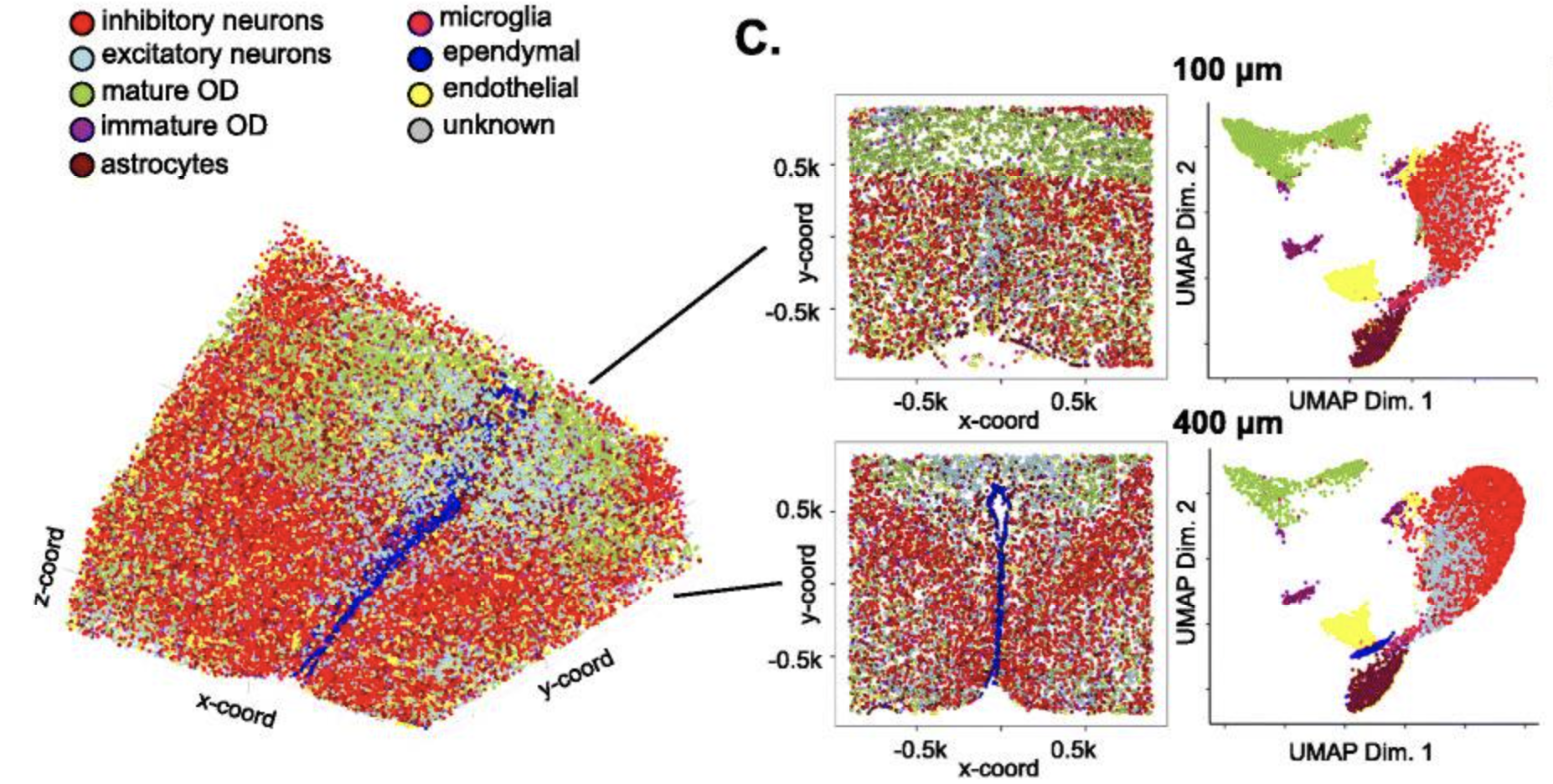
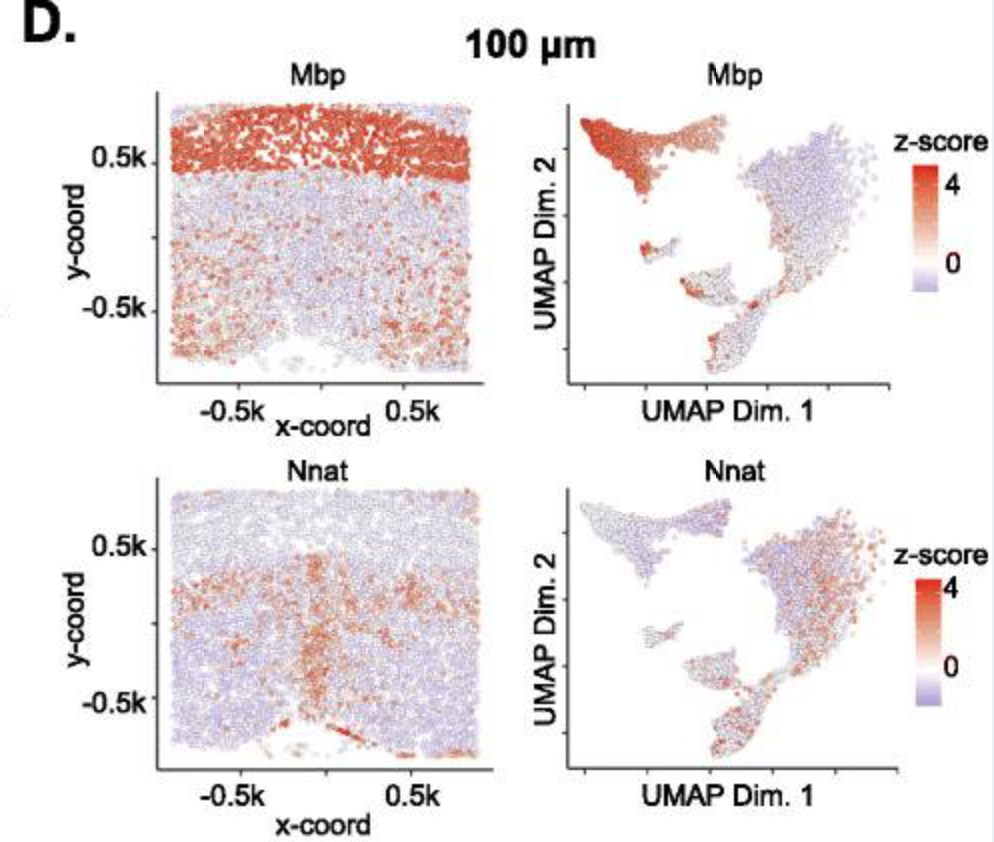
Fig.A Visualization in both expression (top) and physical (bottom) space of the cell types identified by Giotto Analyzer in the pre-optic hypothalamic merFISH dataset, which consists of 12 slices from the same 3D sample (distance unit = 1 μm). Fig.B Heatmap showing the marker genes for the identified cell populations in Fig.A, Fig.C Visualization in both expression and physical space of two representative slices within the z-orientation (100 μm and 400 μm). Fig.D, Fig.E Overlay of gene expression in both expression and physical space for the selected slices in Fig.C, Fig.F.
Click on the images to learn more!